| 1. | Genetic Networks: Large-Scale Mapping of Synthetic Lethal Interactions | |
| a. | Mapping Genetic Interaction Networks | |
| b. | Genetic Interaction Data - Imaging, Processing, and Analysis | |
| c. | Temperature Sensitive Conditional Mutant Collection | |
| d. | Modeling Genetic Networks | |
| 2. | Sigma Deletion Mutant Collection | |
| 3. | High-Content Cell Biology | |
| 4. | Chemical-Genetics | |
| a. | Comparing Chemical-Genetic & Genetic Interaction Networks | |
| b. | Barcoded Yeast Gene Library: Drug-Resistant Mutants and Dosage Suppression | |
| 5. | Mapping Protein-Protein Interaction Networks for Peptide Recognition Modules | |
Mapping Protein-Protein Interactions Networks for Peptide Recognition Modules
People
Raffi Tonikian, Xiaofeng Xin
Protein-protein interactions are of vital importance to the cell as they mediate the assembly of biological complexes and pathways. The fidelity of these interactions in a spatially and temporally controlled fashion is an underlying feature of cellular life; consequently, aberrant interactions often result in abnormal cellular behaviour and disease. An ever-increasing body of data suggests that regulatory proteins are constructed in a modular fashion, with the reiterative and combinatorial use of discrete protein interaction domains. A large subfamily of protein interaction domains is comprised of peptide recognition modules [Pawson and Nash, Science 300: 445-452 (2003)], which are small (50-150 amino acids), independently folding domains that bind linear peptides.
Complete genome sequences have revealed a multitude of these domains, conserved throughout evolution. However, a significant challenge is to generate accurate protein interaction maps linking every peptide recognition module to its binding ligands. The aim of our research project is to define protein interaction networks for metazoan peptide recognition modules, namely for C. elegans SH3 and PDZ domains.
To accomplish this, we are utilizing an integrative approach by combining computational prediction of interactions from phage display ligand consensus sequences with large-scale yeast two-hybrid screening. Phage display [Sidu et al., Curr Opin Chem Biol 7:97-102 (2003)] is a powerful technology for rapidly screening a library of polypeptides that bind to target molecules of interest. The technique takes advantage of the fact that peptides can be displayed on the surfaces of filamentous bacteriophage particles as fusions to the major coat protein. Two-hybrid screening [Fields FEBS J 272: 5391-5399 (2005)] is a molecular biology technique used to discover protein-protein interactions and, with the cloning of the complete orfeome set of genes into the approapriate two-hybrid vectors, it can be applied systematically on a proteome scale [Uetz et al., Nature 403:623-627 (2000); Armstrong et al., Science 303: 540-543 (2004)].
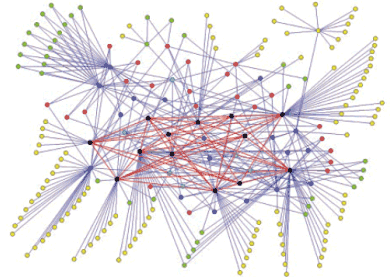
Phage Display SH3 Network
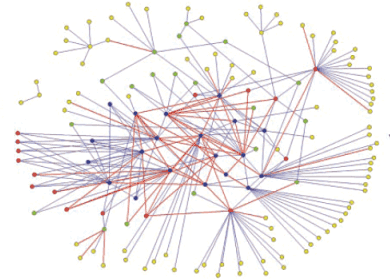
Two Hybrid SH3 Network
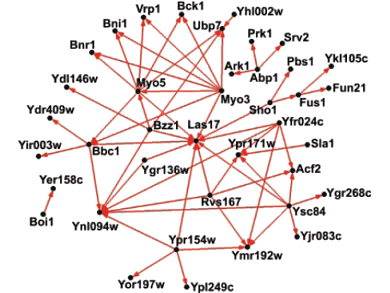
Overlap SH3 Network
Figure 11
To generate an accurate protein interaction network, devoid of false positives, we will determine the intersection between the two independent networks generated by yeast two-hybrid and phage display [Tong et al., Science 295: 321-324 (2002)] (Fig.11). Because of the prominent role of protein interaction domains in regulatory proteins, the high-quality interaction network generated will outline pathways involved in critical cellular events such as the cell cycle, gene expression and signal transduction. Our findings will aid in the understanding of biological processes mediated by regulatory proteins in a developing, multi-cellular organism. These results may ultimately contribute to the development of more effective therapeutics through the targeted disruption of aberrant interactions by small-molecule inhibitors or other synthetic molecules.
back to top

