
Image courtesy of flystuff.com
A crucial aspect of eukaryotic gene regulation occurs at the post-transcriptional level through control of the translation, stability and subcellular localization of mature mRNAs. Post-transcriptional regulation is essential in multiple cell types, notably at synapses in neurons, at the leading edge of migratory cells, in differentiated epithelial cells, and in eggs and early embryos. Defects in post-transcriptional regulation contribute to many human diseases including cancer, cognitive impairment and neurodegenerative disorders. Post-transcriptional events are orchestrated by RNA-binding proteins and non-coding RNAs. We study the mechanisms and functions of post-transcriptional regulation in the fruit fly, Drosophila melanogaster. RNA-binding proteins, their binding sites, as well as the mechanisms through which they regulate their target mRNAs, have been highly conserved in evolution. Our studies in Drosophila will, therefore, help elucidate the mechanisms and functions of post-transcriptional regulation in normal human development as well as disease.
Early development of all animals is programmed by maternal RNAs and proteins that are loaded into the egg by the mother. Thus, gene regulation in the early embryo is post-transcriptional rather than transcriptional, and involves spatial and temporal control of subcellular transcript localization, stability and translation. During the maternal-to-zygotic transition (MZT), a subset of the maternal products is eliminated and zygotic transcription initiates, ultimately leading to the embryo assuming control of its own development (Tadros & Lipshitz 2009, Lipshitz 2015).
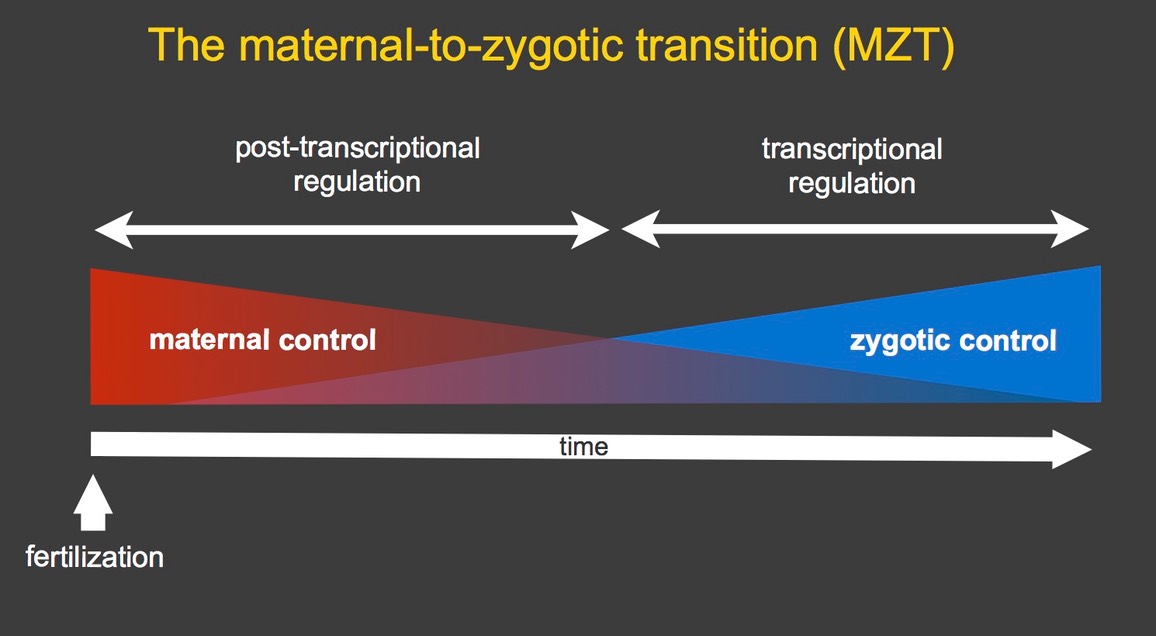
In Drosophila, the MZT occurs during the first few hours after fertilization. We study the mechanisms and functions of post-transcriptional regulation in Drosophila embryos. Using global gene expression profiling we have shown that mRNAs representing over half of all protein coding genes are loaded into the egg by the female fly; after fertilization, a third of these are eliminated during the MZT while two-thirds remain stable (Tadros et al. 2007). Some of the unstable mRNAs are degraded throughout the cytoplasm of the syncytial embryo while others are degraded only on the bulk cytoplasm and are protected from degradation in the germ plasm, which resides at the posterior pole of the embryo and directs germ cell – as distinct from somatic cell – fate (Bashirullah et al. 1999, Siddiqui et al. 2012).
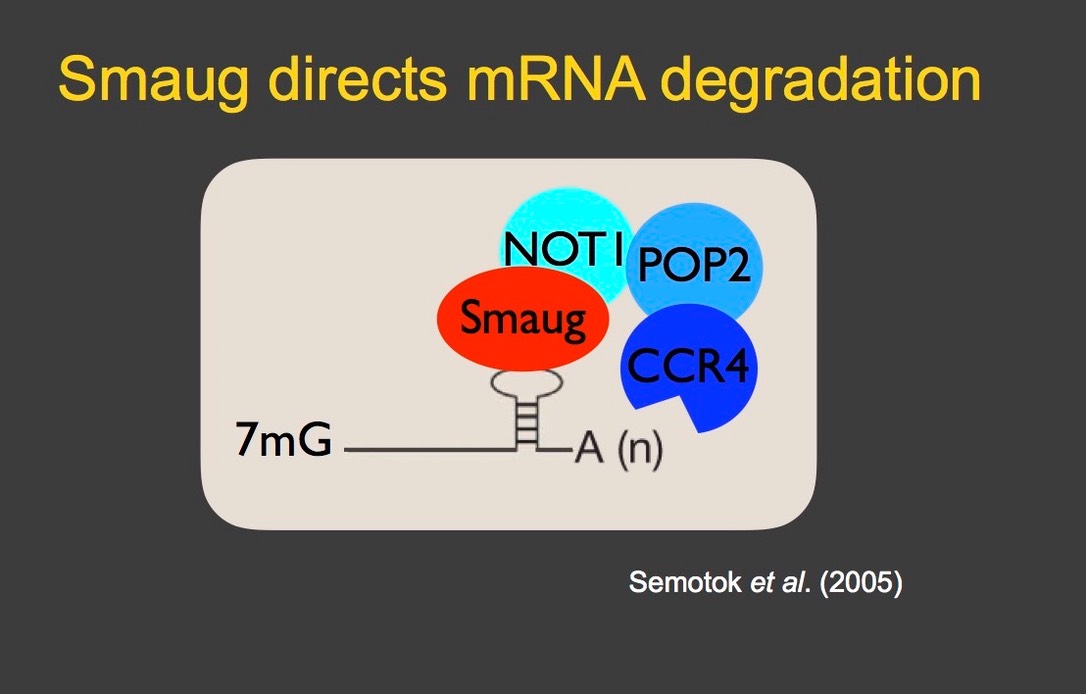
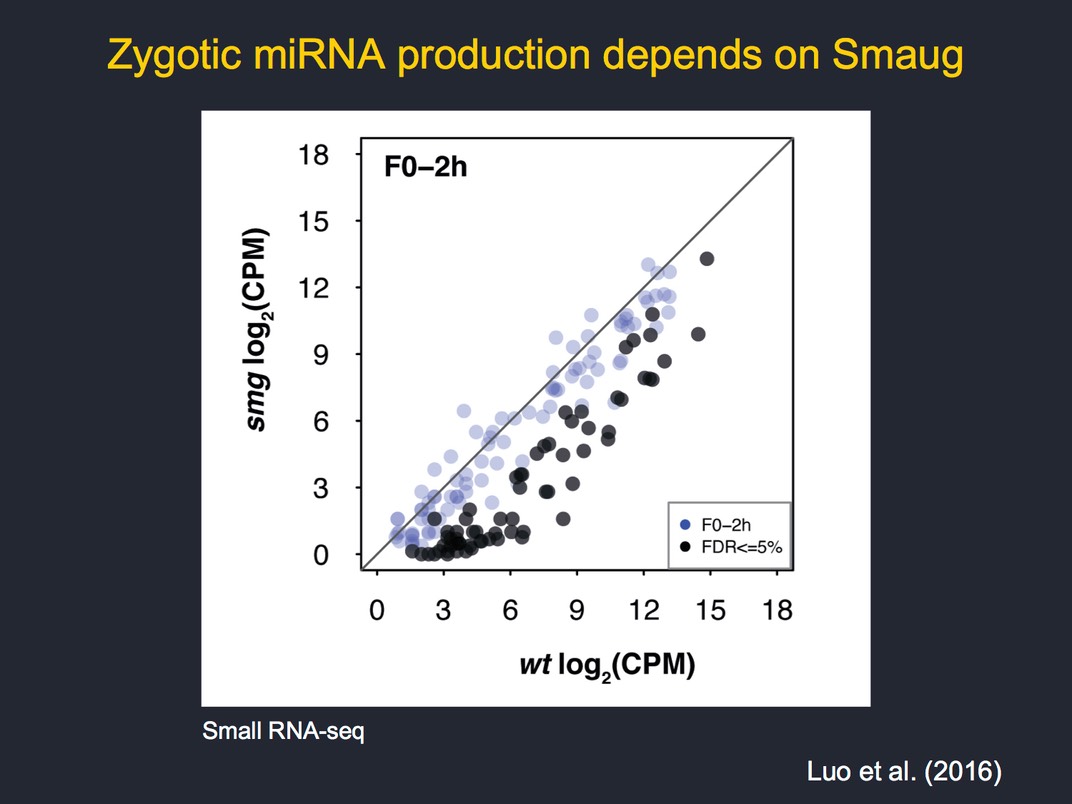
An important and evolutionarily conserved post-transcriptional regulator is the sequence-specific RNA-binding protein, Smaug, which binds hairpin structures called Smaug recognition elements (SREs). Smaug reccruits the CCR4/NOT–deadenylase complex to its target mRNAs to trigger their decay (Semotok et al. 2005). Genome-scale analyses have indicated that Smaug is a major regulator of maternal mRNA decay, being required for elimination of two-thirds of the unstable transcripts. However, not all of the effects of Smaug on transcript stability are direct; our analyses of smaug mutants identified multiple defects in early development, including failure of zygotic genome activation, as an indirect consequence of misregulation of Smaug's target mRNAs (Benoit et al 2009, Luo et al 2016).
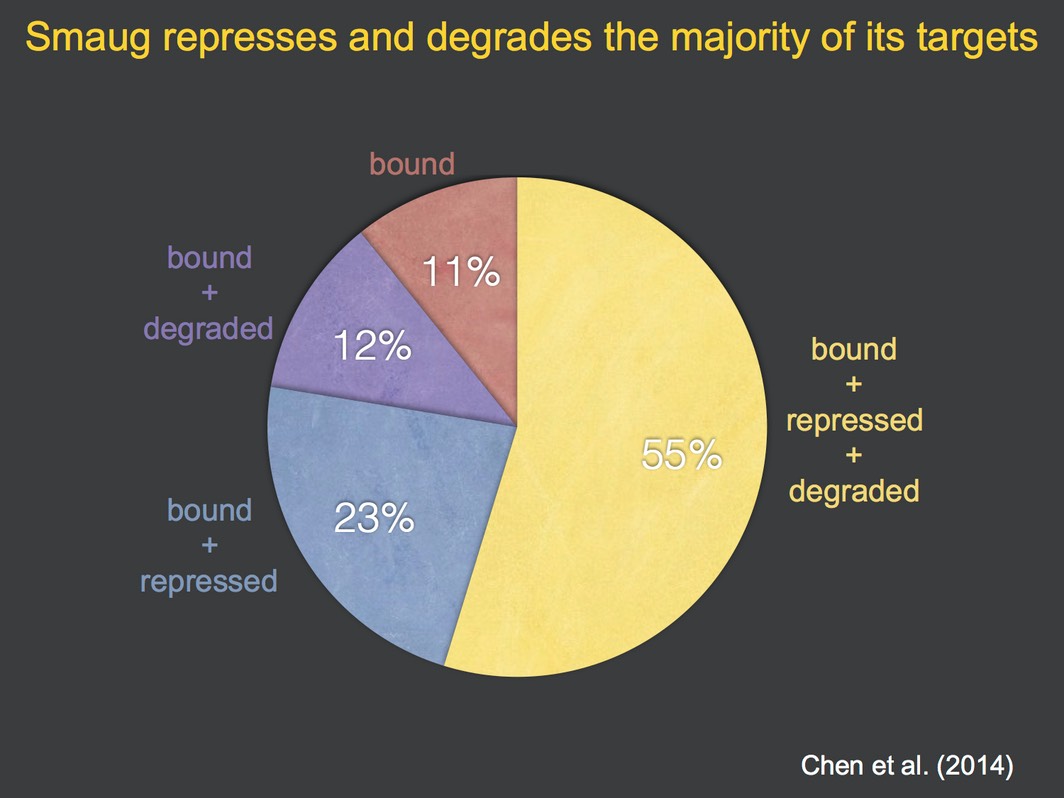
To identify direct targets of Smaug we used antibodies to immunoprecipitate Smaug protein; co-purifying mRNAs were defined using microarrays (‘RIP-Chip’). These studies led to definition of novel cellular and molecular functions for Smaugand showed that the majority of its target mRNAs are both translationally repressed and degraded by Smaug (Chen et al. 2014).
We have established methods for analysis of the hundreds of RNA-binding proteins that are expressed in early embryos. To do so we produce phage-displayed synthetic antibodies for use in immunoprecipitation of these proteins (Laver et al. 2012). We identify co-purifying RNAs by RIP-Chip or next-generation sequencing (‘RIP-Seq’) and associated proteins by mass spectrometry. In parallel we have developed computational methods to identify the binding specificity of RBPs by analyzing the bound mRNAs relative to unbound transcripts using binding-site accessibility as a key criterion (Li et al. 2012, Liet al. 2014).
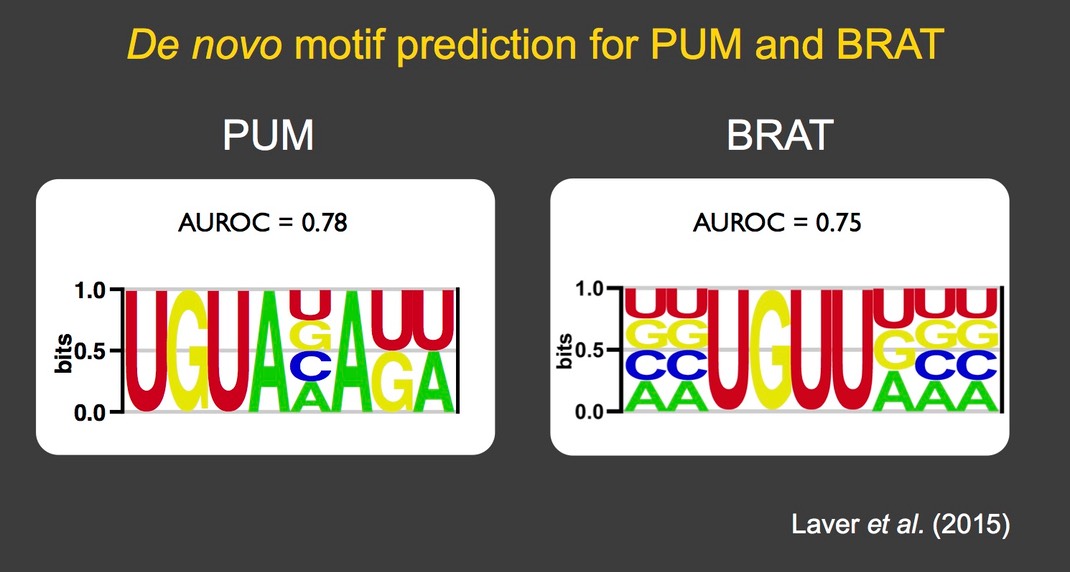
Using these methods, several RNA-binding proteins have been analyzed in detail: Staufen (Laver et al. 2013), Brain tumor and Pumilio (Laver et al. 2015), Rasputin/G3BP (Laver et al. 2020). In so doing we have identified the binding specificity as well as novel biological functions of these proteins, including a major role for Brain tumor in clearance of maternal mRNAs during the MZT and an unexpected role for Rasputin/G3BP in potentiation of mRNA stability and translation.

We developed a high throughput pipeline for the production of synthetic antibodies for the analysis of ribonucleoprotein complexes (Na, Laver et al. 2016). To date we have produced 316 antibodies that recognize 68 ribonucleoprotein complex components. Our long term goal is to produce synthetic antibodies for all 300 RNA-binding proteins that are expressed in early embryos, then to identify the binding specificity, molecular functions and biological roles of these proteins. Over 90% of these are conserved in humans and many are implicated in human disease. Our studies in fruit flies will, therefore, lead to insights into the mechanisms and underlying causes of human diseases that derive from defects in RNA transactions.
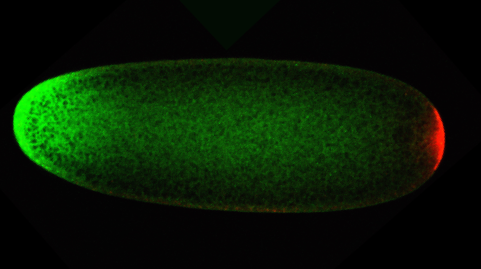
The image shows two RNA-binding proteins in an early Drosophila embryo. An artificial gradient of Smaug (green) has been engineered by localizing the smaug mRNA to the anterior (left) with the bicoid 3’UTR. Endogenous Oskar RNA-binding protein (red) is present only in the germ plasm at the posterior (right). Confocal image by Wael Tadros.