| 1. | Genetic Networks: Large-Scale Mapping of Synthetic Lethal Interactions | |
| a. | Mapping Genetic Interaction Networks | |
| b. | Genetic Interaction Data - Imaging, Processing, and Analysis | |
| c. | Temperature Sensitive Conditional Mutant Collection | |
| d. | Modeling Genetic Networks | |
| 2. | Sigma Deletion Mutant Collection | |
| 3. | High-Content Cell Biology | |
| 4. | Chemical-Genetics | |
| a. | Comparing Chemical-Genetic & Genetic Interaction Networks | |
| b. | Barcoded Yeast Gene Library: Drug-Resistant Mutants and Dosage Suppression | |
| 5. | Mapping Protein-Protein Interaction Networks for Peptide Recognition Modules | |
Chemical-Genetics:
Comparing Chemical-Genetic & Genetic Interaction Networks
People
Andres Lopez, Ainslie Parsons (Lab Alumnus)
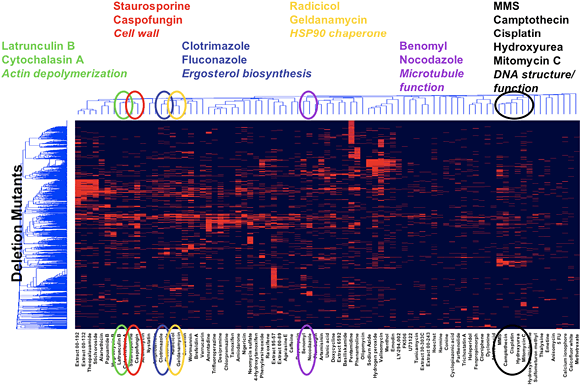
Genomic approaches in yeast have pioneered a whole new era in biology and they have enable technologies with a direct application to the field of natural/synthetic chemical products. We are applying yeast chemical-genetics approaches to gain insights into the mechanism of drug action. Perhaps the most powerful approach is the chemical-genetic information obtained from scoring each of the ~5000 viable deletion mutant strains for sensitivity to a specific compound using a parallel growth assay and barcode microarray readout.
Each mutant carries a unique 20 base pair oligonucleotide barcode, which upon amplification and hybridization to a barcode microarray provides information on the abundance of the mutant in a particular growth assay [Giaever et al., Nature 418:387-391 (2002)]. Thus, with a small volume of liquid growth medium, all ~5000 deletion mutants can be scored for fitness in the presence of a bioactive active compound.
Gene deletions that render cells hypersensitive to a specific compound identify pathways that buffer the cell against the toxic effects of the compound. We created a detailed compendium of chemical-genetic interaction profiles, the set of viable gene deletion mutants that are hypersensitive to a particular compound, and found that clustering of the results organizes yeast genes into functional pathways but also organizes compounds into related sets with similar biological functions [Parsons et al., Cell 126:611-625 (2006)] (Fig.6). Thus, chemical-genetic profiling with yeast has proven to be a powerful new system for determining the mode-of-action of bioactive molecules.
A
B
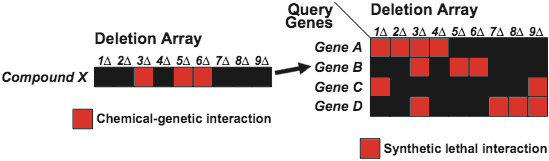
Figure 7
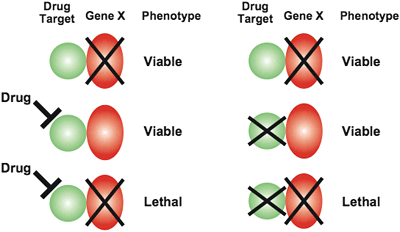
Because a deletion mutant provides a good model for the physiological effects of an inhibitory compound, the yeast synthetic lethal genetic interaction map provides a key for interpretation of the chemical-genetic profile of a compound [Parsons et al., Nature Biotechnology 22, 62 - 69 (2004)] (Fig.7).
In a proof-of-principle study, we've shown that compounds whose chemical-sensitivity profile resemble the synthetic lethal genetic interaction profile of a specific query gene may very well target the product of the identified gene or its corresponding pathway. We are comparing our synthetic lethal and chemical-genetic interaction networks computationally and predicting the targets pathways of specific compounds.