Project Links
| 1. | Genetic Networks: Large-Scale Mapping of Synthetic Lethal Interactions | |
| a. | Mapping Genetic Interaction Networks | |
| b. | Genetic Interaction Data - Imaging, Processing, and Analysis | |
| c. | Temperature Sensitive Conditional Mutant Collection | |
| d. | Modeling Genetic Networks | |
| 2. | Sigma Deletion Mutant Collection | |
| 3. | High-Content Cell Biology | |
| 4. | Chemical-Genetics | |
| a. | Comparing Chemical-Genetic & Genetic Interaction Networks | |
| b. | Barcoded Yeast Gene Library: Drug-Resistant Mutants and Dosage Suppression | |
| 5. | Mapping Protein-Protein Interaction Networks for Peptide Recognition Modules | |
Genetic Networks:
Genetic Interaction Data - Imaging, Processing and Analysis
Collaborative Project with Brenda Andrews' Lab
People
Huiming Ding, Toufighi, Judice Koh
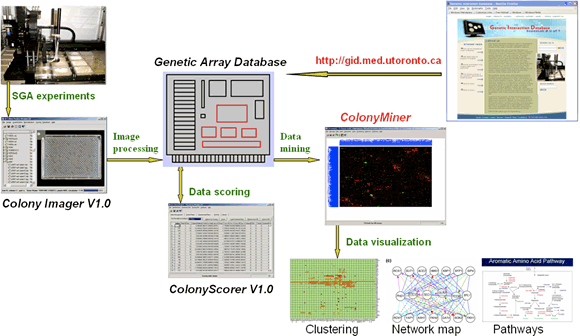
In SGA, identification of genetic interactions is based on double mutant fitness which is assessed as a measure of colony size. In order to meet our throughput demands, we have designed and developed an automated, quantitative approach to score fitness. This computational platform facilitates large-scale SGA data analysis and provides an analysis pipeline consisting of the following steps: image processing, fitness scoring, data storage, statistical analysis and data mining (Fig.4).
Several laboratories around the world have implemented this system and actively contribute data to our growing repository, the Genetic Interaction Database (GID).