| 1. | Genetic Networks: Large-Scale Mapping of Synthetic Lethal Interactions | |
| a. | Mapping Genetic Interaction Networks | |
| b. | Genetic Interaction Data - Imaging, Processing, and Analysis | |
| c. | Temperature Sensitive Conditional Mutant Collection | |
| d. | Modeling Genetic Networks | |
| 2. | Sigma Deletion Mutant Collection | |
| 3. | High-Content Cell Biology | |
| 4. | Chemical-Genetics | |
| a. | Comparing Chemical-Genetic & Genetic Interaction Networks | |
| b. | Barcoded Yeast Gene Library: Drug-Resistant Mutants and Dosage Suppression | |
| 5. | Mapping Protein-Protein Interaction Networks for Peptide Recognition Modules | |
Chemical-Genetics:
Barcoded Yeast Gene Library: Drug-Resistant Mutants and Dosage Suppression
People
Cheuk-Hei Ho, Leslie Magtanong, Sarah Barker, Kusala Jayasuriya
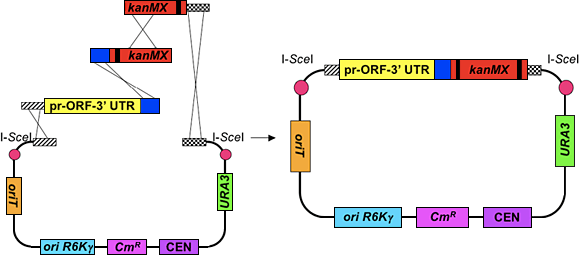
We developed a systematic, high-throughput method to clone all of the Saccharomyces cerevisiae genes under the control of their endogenous regulatory regions into barcoded vectors in order to obtain a genome-wide library of CEN-based (low copy) clones (Fig. 8). We will then use a bacterial conjugation technique called MAGIC (mating-assisted genetically integrated cloning) [Li and Elledge Nat. Gen. 37 :311-319 (2005)] to convert the CEN-based clones into two-micron (high-copy) clones.
The libraries we are building will complement current overexpression libraries that have been described in the literature, including systematic GAL1 -ORF libraries [Gelperin et al., Genes Dev. 19 :2816-2826 (2005); Sopko et al. Mol. Cell 21 :319-330 (2006)]. Each of our clones contains a single ORF flanked by ~800 bp and ³ 150 bp of upstream and downstream sequence respectively; thus, the expression of each ORF is controlled, to some extent, by its own promoter and 3' UTR. Each ORF is barcoded with its unique kanMX cassette from the corresponding gene deletion strain [Shoemaker et al. Nat. Gen . 14 :450-456 (1996)]. By including the barcodes in our library, our clones can be used accordingly in barcode microarray experiments.
We will apply the use of the single copy library towards the cloning and characterization of drug-resistant mutants, which often identify the target if the drug. We also plan to use the high copy library for identifying genes that led to dosage resistance to a particular compound or drug, which can also identify the target of the drug [Parsons et al., Cell 126:611-625 (2006)].
In addition, the high copy library can be used to score genetic interactions. Specifically, we will use the high copy library for dosage suppression genetic interaction analysis, suppressing the lethality associated with temperature sensitive conditional mutants. This analysis will generate a new type of edge on our genetic interaction networks, which are largely composed of synthetic lethal interactions, and should allow us to begin to order pathway components by combining overexpression and deletion mutant perturbations.
back to top