Publications
2012
Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells.
Rugg-Gunn PJ, Cox BJ, Lanner F, Sharma P, Ignatchenko V, McDonald AC, Garner J, Gramolini AO, Rossant J, Kislinger T.
Dev Cell. 2012 Apr 17;22(4):887-901. Epub 2012 Mar 15.
PubMed PMID: 22424930
PubMed Central PMCID: PMC3405530
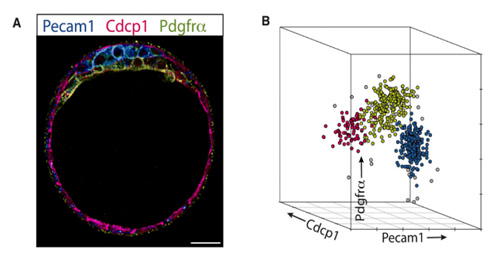
Immuno-florescent confocal image of a 4.5 day old mouse embryo with cell surface markers for each of the three cell fate lineages (A, epiblast; blue, Pecam1; trophoblast, red; Cdcp1, primitive endoderm; green, Pdgfrα). These same markers are used to separate and isolate the different cell types from embryos using flow cytometry (B colours correspond to markers in A). The isolation of single cells enables the monitoring of cell fate development and population dynamics over time.
2011
Integrated microarray and ChIP analysis identifies multiple Foxa2 dependent target genes in the notochord.
Tamplin OJ, Cox BJ, Rossant J.
Dev Biol. 2011 Dec 15;360(2):415-25. Epub 2011 Oct 8.
PubMed PMID: 22008794
Translational analysis of mouse and human placental protein and mRNA reveals distinct molecular pathologies in human preeclampsia.
Cox B, Sharma P, Evangelou AI, Whiteley K, Ignatchenko V, Ignatchenko A, Baczyk D, Czikk M, Kingdom J, Rossant J, Gramolini AO, Adamson SL, Kislinger T.
Mol Cell Proteomics. 2011 Dec;10(12):M111.012526. Epub 2011 Oct 10.
PubMed PMID: 21986993
PubMed Central PMCID: PMC3237089
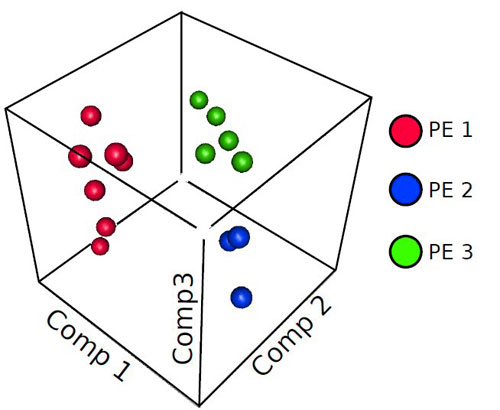
A three dimensional plot of the top three principal components that separate different molecular classes of preeclampsia. This analysis is the first to show that there are distinct molecular expression profiles among patients with clinically similar pathology. These results imply that the treatment of preeclampsia will require the development of therapies tailored to different molecular causes. This is analogous to the treatment of other multi-factorial diseases, such as cancer.
Porcupine homolog is required for canonical Wnt signaling and gastrulation in mouse embryos.
Biechele S, Cox BJ, Rossant J.
Dev Biol. 2011 Jul 15;355(2):275-85. Epub 2011 Apr 30.
PubMed PMID: 21554866
Back to top2010
Phenotypic annotation of the mouse X chromosome.
Cox BJ, Vollmer M, Tamplin O, Lu M, Biechele S, Gertsenstein M, van Campenhout C, Floss T, Kühn R, Wurst W, Lickert H, Rossant J.
Genome Res. 2010 Aug;20(8):1154-64. Epub 2010 Jun 14.
PubMed PMID: 20548051
PubMed Central PMCID: PMC2909578
X-linked gene database
Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo.
Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J.
Proc Natl Acad Sci U S A. 2010 Jun 15;107(24):10783-90. Epub 2010 May 17.
PubMed PMID: 20479220
PubMed Central PMCID: PMC2890770
Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2.
Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J.
Development. 2010 Feb;137(3):395-403.
PubMed PMID: 20081188
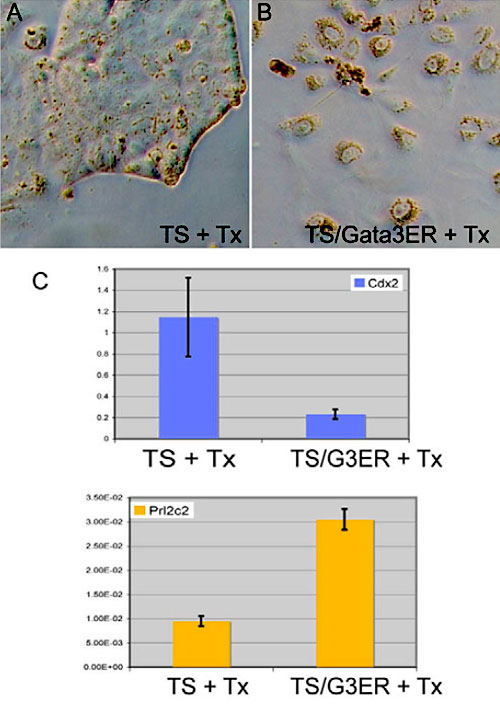
The role of the transcription factor Gata3 in trophoblast development. Increased expression of Gata3 causes trophoblast stem cells (A) to differentiated in to trophoblast giant cells (B). This is confirmed by the loss of expression of Cdx2©, a trophoblast stem cell marker gene, and an increase in expression of Prl2c2©, a marker of differentiated trophoblast cells. Gata3 with inducible activity was expressed in trophoblast cells and induced with tamoxifen.
The study identifies Gata3 as having a dual role in trophoblast stem cell maintenance and differentiation, as well it identified Gata3 as functioning in parallel to the other known regulator of trophoblast development, Cdx2.
2009
Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology.
Cox B, Kotlyar M, Evangelou AI, Ignatchenko V, Ignatchenko A, Whiteley K, Jurisica I, Adamson SL, Rossant J, Kislinger T.
Mol Syst Biol. 2009;5:279. Epub 2009 Jun 16.
PubMed PMID: 19536202
PubMed Central PMCID: PMC2710868
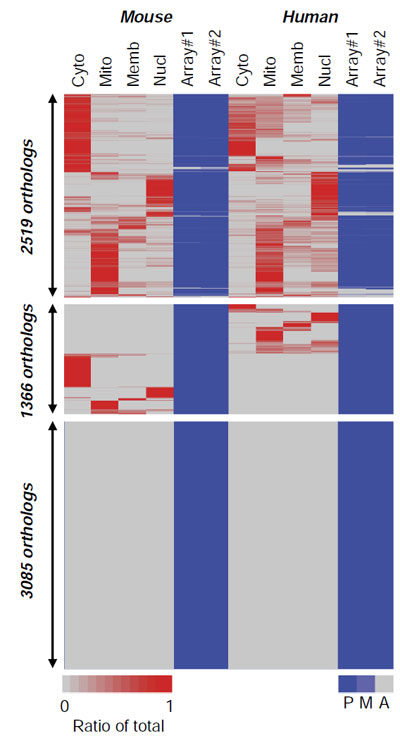
Heat map of the integrated protein and mRNA expression profiles of human and mouse placentas. The proteins are separated into different subcellular fractions (top labels: cyto, cytoplasm; Mito, mitochondria; Memb, membrane; Nucl, nucleus), replicate DNA microarrays were used to assess mRNA expression. This analysis is limited to the ~11,000 1:1 orthologs between the two species.
The findings of this analysis and others in the study show that while human and mouse have many genes expressed in common there is significant unique gene expression. This suggests that there are limits to the ability of mice to model human pathologies.
2008
Microarray analysis of Foxa2 mutant mouse embryos reveals novel gene expression and inductive roles for the gastrula organizer and its derivatives.
Tamplin OJ, Kinzel D, Cox BJ, Bell CE, Rossant J, Lickert H.
BMC Genomics. 2008 Oct 30;9:511.
PubMed PMID: 18973680
PubMed Central PMCID: PMC2605479
Dynamic expression of Lrp2 pathway members reveals progressive epithelial differentiation of primitive endoderm in mouse blastocyst.
Gerbe F, Cox B, Rossant J, Chazaud C.
Dev Biol. 2008 Jan 15;313(2):594-602. Epub 2007 Nov 12.
PubMed PMID: 18083160
Back to top2007
Integrated proteomic and transcriptomic profiling of mouse lung development and Nmyc target genes.
Cox B, Kislinger T, Wigle DA, Kannan A, Brown K, Okubo T, Hogan B, Jurisica I, Frey B, Rossant J, Emili A.
Mol Syst Biol. 2007;3:109. Epub 2007 May 8.
PubMed PMID: 17486137
PubMed Central PMCID: PMC2673710
Trophoblast-specific gene manipulation using lentivirus-based vectors. Biotechniques.
Georgiades P, Cox B, Gertsenstein M, Chawengsaksophak K, Rossant J.
Biotechniques 2007 Mar;42(3):317-8, 320, 322-5.
PubMed PMID: 17390538
Back to top2006
Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling.
Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A.
Cell. 2006 Apr 7;125(1):173-86.
PubMed PMID: 16615898
Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics.
Cox B, Emili A.
Nat Protoc. 2006;1(4):1872-8.
PubMed PMID: 17487171
Back to top2005
Dissecting Wnt/beta-catenin signaling during gastrulation using RNA interference in mouse embryos.
Lickert H, Cox B, Wehrle C, Taketo MM, Kemler R, Rossant J.
Development. 2005 Jun;132(11):2599-609. Epub 2005 Apr 27.
PubMed PMID: 15857914
Integrating gene and protein expression data: pattern analysis and profile mining.
Cox B, Kislinger T, Emili A.
Methods. 2005 Mar;35(3):303-14. Epub 2005 Jan 12. Review.
PubMed PMID: 15722226
Back to top2003
PRISM, a generic large scale proteomic investigation strategy for mammals.
Kislinger T, Rahman K, Radulovic D, Cox B, Rossant J, Emili A.
Mol Cell Proteomics. 2003 Feb;2(2):96-106. Epub 2003 Feb 10.
PubMed PMID: 12644571
Back to top