


WHAT WE DO
Our lab is interested in uncovering cancer signal transduction networks and elucidating mechanisms of their regulation. We are currently involved in a project to comprehensively map the genetic susceptibilities of serous ovarian cancer using functional genetic screens.
Interested and motivated postdoctoral fellows are highly encouraged to contact the lab and graduate students can apply through the Department of Medical Biophysics or the Department of Immunology at the University of Toronto.
RECENT PUBLICATIONS

Go with the flow: GEF-H1 mediated shear stress mechanotransduction in neutrophils.
Nov 2017
Small GTPase
Neutrophils in circulation experience significant shear forces due to blood flow when they tether to the vascular endothelium. Neutrophil mechanotransduction responses occur through mechanisms that are not yet fully understood. Here, we propose that GEF-H1-dependent cell spreading and crawling in shear stress-dependent neutrophil recruitment from the vasculature are due to the specific localization of Rho-induced contractility in the uropod.
More

MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity.
Oct 2017
Science Signaling
ARHGEF2 is a multifunctional guanine exchange factor involved in RhoA activation and MAPK pathway activation through its noncatalytic scaffold function. Here, we have uncovered a previously unknown phosphoregulatory switch, which regulates the subcellular localization and activity of ARHGEF2 that couples microtubules to the actin cytoskeleton to establish epithelial cell polarity.
More

Co-dependency between KRAS addiction and ARHGEF2 promotes an adaptive escape from MAPK pathway inhibition.
June 2017
Small GTPase
Oncogenic KRAS engages multiple effector pathways including the MAPK cascade to promote proliferation and survival of pancreatic cancer cells. In this study, we investigate the relationship between KRAS and ARGEF2, and further illustrate that ARHGEF2 expression may increase the efficacy of MAPK inhibitors for treatment of RAS-dependent pancreatic cancers.
More

Ubiquitin ligase RNF146 coordinates bone dynamics and energy metabolism.
June 2017
The Journal of Clinical Investigation
This study shows RNF146 is required to coordinate β-catenin signaling within the osteoblast lineage during embryonic and postnatal bone development.
More

RANKL coordinates multiple osteoclastogenic pathways by regulating expression of ubiquitin ligase RNF146.
April 2017
The Journal of Clinical Investigation
This study shows that RANKL-mediated suppression of RNF146 results in the stabilization of its substrates, thus acts as an inhibitory switch to control osteoclastogenesis and cytokine production and may be a control point underlying the pathogenesis of chronic inflammatory diseases.
More

Interrogation of Functional Cell-Surface Markers Identifies CD151 Dependency in High-Grade Serous Ovarian Cancer.
March 2017
Cell Reports
This study reveals cell-surface vulnerabilities associated with high grade serous carcinoma (HGSC), provides a framework for identifying therapeutic targets, and reports a role for tetraspanin CD151 in HGSC.
More
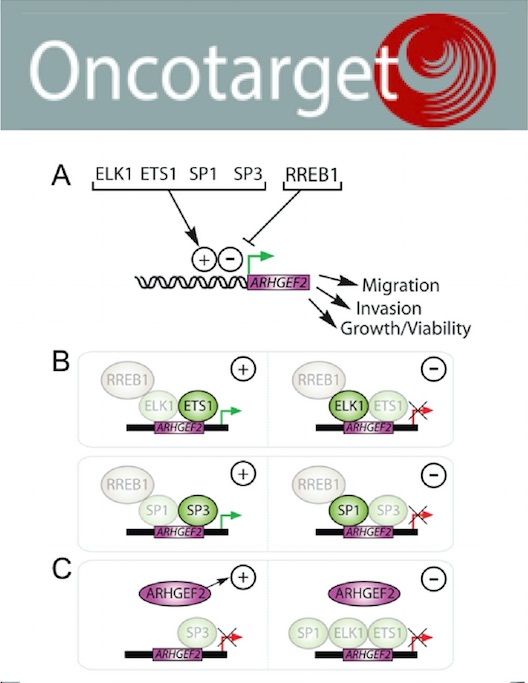
An oncogenic KRAS transcription program activates the RHOGEF ARHGEF2 to mediate transformed phenotypes in pancreatic cancer.
January 2017
Oncotarget
Our results identify that oncogenic KRAS activates ARHGEF2 through a minimal RAS responsive promoter and that this transcription factor program is required for RAS transformation and provides mechanistic insight into the highly metastatic behavior of pancreatic cancer.
More

Reciprocal stabilization of ABL and TAZ regulates osteoblastogenesis through transcription factor RUNX2.
December 2016
The Journal of Clinical Investigation
Our study demonstrates an interplay between ABL and TAZ that controls the mesenchymal maturation program toward the osteoblast lineage and is mechanistically distinct from the established model of lineage-specific maturation.
More

GEF-H1 is necessary for neutrophil shear stress–induced migration during inflammation.
October 2016
The Journal of Cell Biology
Our results identify GEF-H1 as a component of the shear stress response machinery in neutrophils required for a fully competent immune response to bacterial infection.
More
Transcriptional Regulation of miR-31 by Oncogenic KRAS Mediates Metastatic Phenotypes by Repressing RASA1.
March 2016
Molecular Cancer Research
In the study miR-31 is identified as a RAS effector that enhances invasion-migration of pancreatic cancer cells via downregulation of the miR-31 target gene RASA1 and activation of RhoA. Expression of miR-31 is coupled to the expression of oncogenic KRAS and activity of the MAPK pathway.
More

Mechanistic insight into GPCR-mediated activation of the microtubule-associated RhoA exchange factor GEF-H1.
September 2014
Nature Communications
The coordinated displacement of GEF-H1 from microtubules by G-proteins and its dephosphorylation by PP2A demonstrate a multistep GEF-H1 activation and present a unique mechanism coupling GPCR signalling to Rho activation.
More

The RhoGEF GEF-H1 Is Required for Oncogenic RAS Signaling via KSR-1.
February 2014
Cancer Cell
Our results identify GEF-H1 as an amplifier of MAPK signaling and provide mechanistic insight into the progression of RAS mutant tumors.
More

The 3BP2 Adapter Protein Is Required for Chemoattractant-Mediated Neutrophil Activation.
July 2012
Journal of Immunology
Our results reveal an obligate requirement for the adapter protein 3BP2 in G protein-coupled receptor-mediated neutrophil function.
More
Mechanistic insight into the microtubule and actin cytoskeleton coupling through dynein-dependent RhoGEF inhibition.
March 2012
Molecular Cell
These studies demonstrate a pivotal role of Tctex-1 as a negative regulator of actin filament organization through its control of Lfc in the crosstalk between microtubule and actin cytoskeletons.
More

Essential gene profiles in breast, pancreatic, and ovarian cancer cells.
February 2012
Cancer Discovery
This study presents a resource of genome-scale, pooled shRNA screens for 72 breast, pancreatic, and ovarian cancer cell lines that will serve as a functional complement to genomics data, facilitate construction of essential gene profiles, help uncover synthetic lethal relationships, and identify uncharacterized genetic vulnerabilities in these tumor types.
More

Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism.
December 2011
Cell
Here we show that Tankyrase, a member of the poly(ADP-ribose)polymerase (PARP) family, regulates 3BP2 stability through ADP-ribosylation and subsequent ubiquitylation by the E3-ubiquitin ligase RNF146 in osteoclasts.
More

Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease
December 2011
Cell
Here we use a Tankyrase recognition motif to rationalize all known Tankyrase substrates and explains the basis for cherubism-causing mutations in the Tankyrase substrate 3BP2. Structural and sequence information allows us to also predict and validate other Tankyrase targets, including Disc1, Striatin, Fat4, RAD54, BCR, and MERIT40.
More

3BP2-deficient mice are osteoporotic with impaired osteoblast and osteoclast functions
August 2011
Journal of Clinical Investigation
These findings reveal an unanticipated role for the 3BP2 adapter protein in osteoblast function and in coordinating bone homeostatic signals in both osteoclast and osteoblast lineages.
More