Our research focuses on mechanisms underlying the regulation of gene expression and how these mechanisms are disrupted in human diseases and disorders. Most of our research is directed at understanding how alternative splicing is regulated and integrated with other layers of gene expression to control fundamental biological processes. For example, we have discovered alternative splicing "switches" with critical roles in the regulation of transcriptional programs required for neural and embryonic cell fate. More recently, we have discovered a highly conserved alternative splicing regulatory network involving tiny (3-27 nt) neuronal microexons, and have provided evidence that the disruption of this network represents a common mechanism underlying autism spectrum disorders. Our research encompasses a wide range of approaches, from bioinformatics and functional genomics to focused molecular, biochemical, cell biological methods, as well as the generation of animal models. We have pioneered the development and application of technologies for the genome-wide quantitative profiling of transcriptomes, RNA interactomes, as well as new CRISPR-based screens designed to comprehensively define RNA regulatory networks. These efforts are uncovering remarkable landscapes of new regulation that await further investigation. They are also providing insight into new therapeutic strategies for human diseases and disorders.
Selected Review and Perspective Articles

Blencowe, B.J (2017) The Relationship between Alternative Splicing and Proteomic Complexity. Trends Biochem Sci.42:407-408.
[PUBMED] [PDF]
|

Blencowe, B.J. (2015). Reflections for the 20th anniversary issue of RNA journal. RNA. 21(4): 573-575
[PUBMED] [PDF]
|
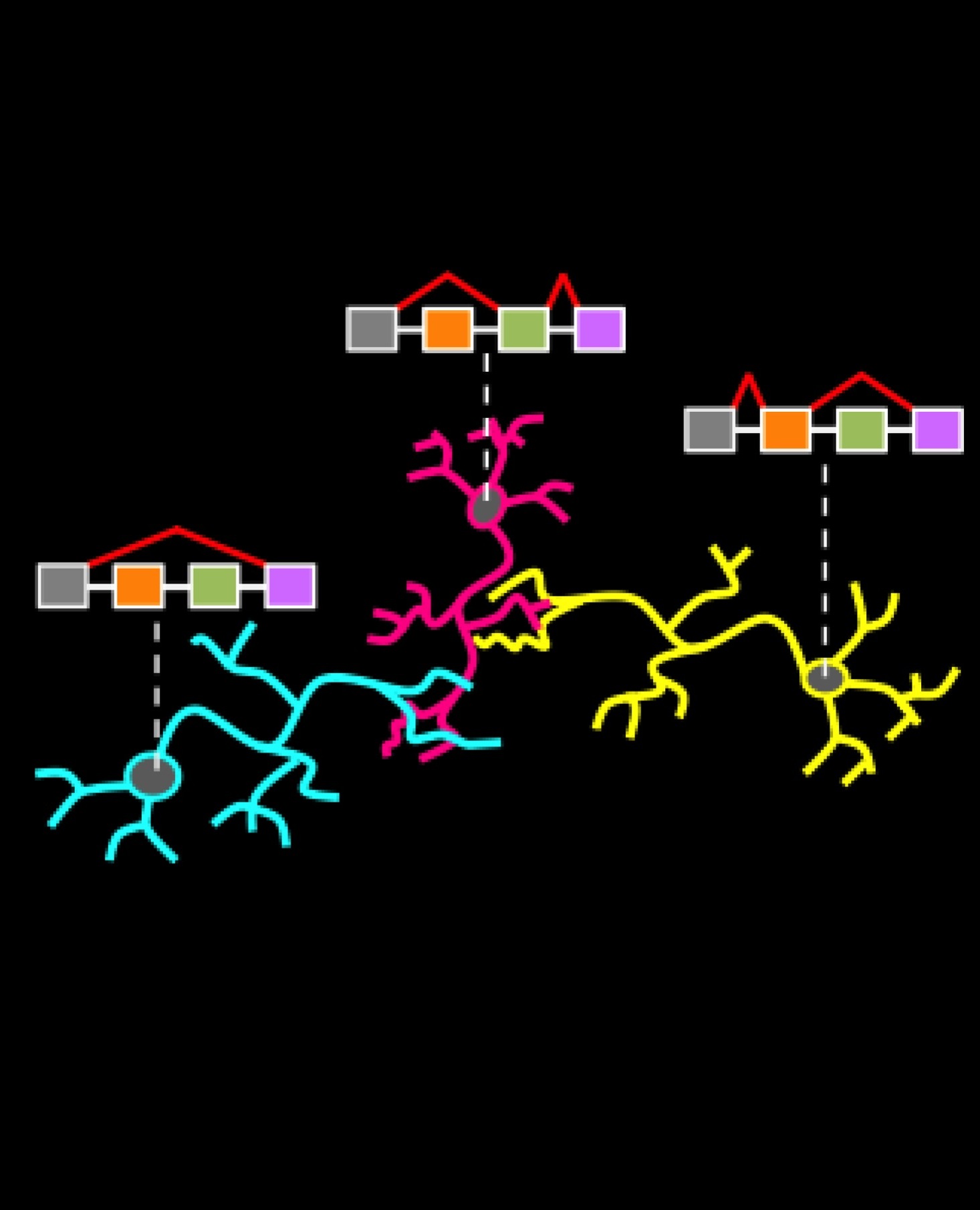
Raj, B. and Blencowe, B.J (2015) Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles, Neuron, 81(1):14-27.
[PUBMED] [PDF]
|

Braunschweig, U., Gueroussov, S., Plocik, A.M., Graveley, B.R. and Blencowe, B.J (2013) Dynamic integration of Splicing within Gene Regulatory Pathways, Cell, 152:1252-1269.
[PUBMED] [PDF]
|
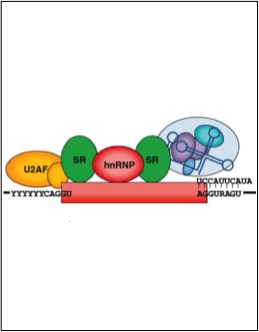
Irimia, M, Blencowe, B.J. (2012) Alternative splicing: decoding an expansive regulatory layer, Current Opinion in Cell Biology, 24: 3.
[PUBMED] [PDF]
|
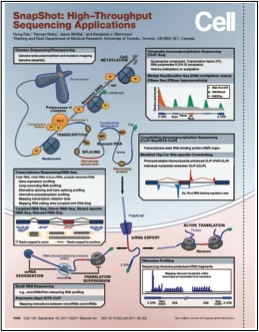
Han, H., Nutiu, R., Moffat, J. and Blencowe, B.J. (2011) SnapShot: High Throughput Sequencing Applications, Cell 146(6):1044-1044.e2.
[PUBMED] [PDF] |
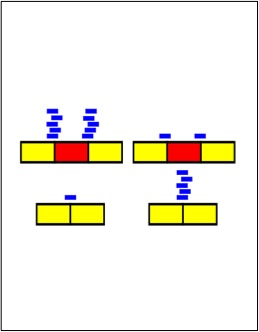
Blencowe, B.J., Ahmad, S. and Lee,L.J.(2009) Current generation high-throughput sequencing: deepening insights into mammalian transcriptomes, Genes and Development 23:1379-1386.
[PUBMED] [PDF]
|
Blencowe, B.J. (2006) Alternative Splicing: New insights from global analyses, Cell 126:37-47.
[PUBMED] [PDF] |
Current Research Topics and Selected Publications
Deep surveying of transcriptome complexity, global mapping of RNA-RNA interactions and deciphering RNA regulatory codes
Weatheritt, R.J., Sterne-Weiler, T. and Blencowe, B.J (2016) The ribosome-engaged landscape of alternative splicing. Nat Struct Mol Biol. 23:1117-1123.
[PUBMED] [PDF]
|

Sharma, E., Sterne-Weiler, T., O'Hanlon, D. and Blencowe, B.J. (2016). Global Mapping of Human RNA-RNA Interactions. Mol. Cell. 62:618-626.
[PUBMED] [PDF]
-Highlighted in Molecular Cell and TIBS
|
Xiong, H.Y., Alipanahi, B., Lee, L.J., Bretschneider, H., Merico, D., Yuen, R.K., Hua, Y., Gueroussov, S., Najafabadi, H.S., Hughes, T.R., Morris, Q., Barash, Y., Krainer, A.R., Jojic, N., Scherer, S.W., Blencowe, B.J. and Frey, B.J. (2015). The human splicing code reveals new insights into the genetic determinants of disease. Science. 347(6218):12.
[PUBMED] [PDF]
-Highlighted in Science Perspectives
-Featured in several media reports including The Globe and Mail and The Varsity.
-Recommended by Faculty of 1000
|
Braunschweig, U., Barbosa-Morais, N.L., Pan, Q., Nachman, E., Alipanahi, B., Gonatopoulos-Pourtnazis, T., Frey, B., Irimia, M. & Blencowe, B.J. (2014). Widespread intron retention in mammals functionally tunes transcriptomes. Genome Research, 24(11): 1774-1786.
[PUBMED] [PDF]
-Highlighted in Nature Reviews Genetics article
|

Barash, Y., Calarco, J.A., Gao, W., Pan, Q., Wang, X., Shai, O., Blencowe, B.J., and Frey, B.J. (2010). Deciphering the splicing code. Nature, 465: 53-59.
[PUBMED] [PDF]
-Featured in Nature Editorials, in several News and Views articles in Nature, Nature Biotech, Cell Res, and numerous media reports including CBC,and Toronto Star
-Featured on the journal cover
-Recommended by Faculty of 1000
-Featured in a Nature Biotech article "Trends in Computational Biology-2010"
|

Pan, Q., Shai, O., Lee, L., Frey, B.J., and Blencowe, B.J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genetics. 40:1413-1415.
[PUBMED] [PDF]
-Highlighted in Nature and Nature Genetics Editorial, Research Highlight of Nature Review Genetics.
-Highlighted on the journal cover
|
|
Discovery and characterization of splicing regulators and exon networks with critical roles in development
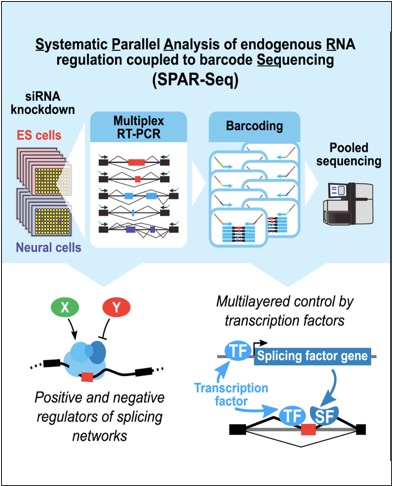
Han, H., Braunschweig, U., Gonatopoulos-Pournatzis, T., Weatheritt, R.J., Hirsch, C.J., Ha, K.C., Radovani, E., Sterne-Weiler, T., Wang, J., O’Hanlon, D., Pan, Q., Ray, D., Vizeacoumar, F., Datti, A., Magomedova, L., Cummins, L.C., Hughes, T.R., Greenblatt, J.F., Wrana, J.L., Moffat, J. and Blencowe, B.J. (2017).
Multilayered control of alternative splicing regulatory networks by transcription factors. Mol. Cell 65: 539-55.
[PUBMED] [PDF]
|
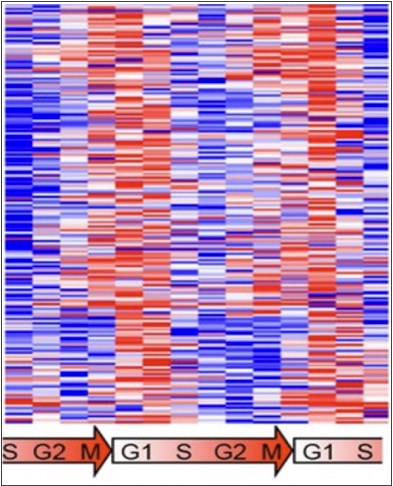
Dominguez, D., Tsai, Y.H., Weatheritt, R., Wang, Y., Blencowe, B.J. and Wang, Z. (2016) An extensive program of periodic alternative splicing linked to cell cycle progression. eLife 5:e10288.
[PUBMED] [PDF]
|
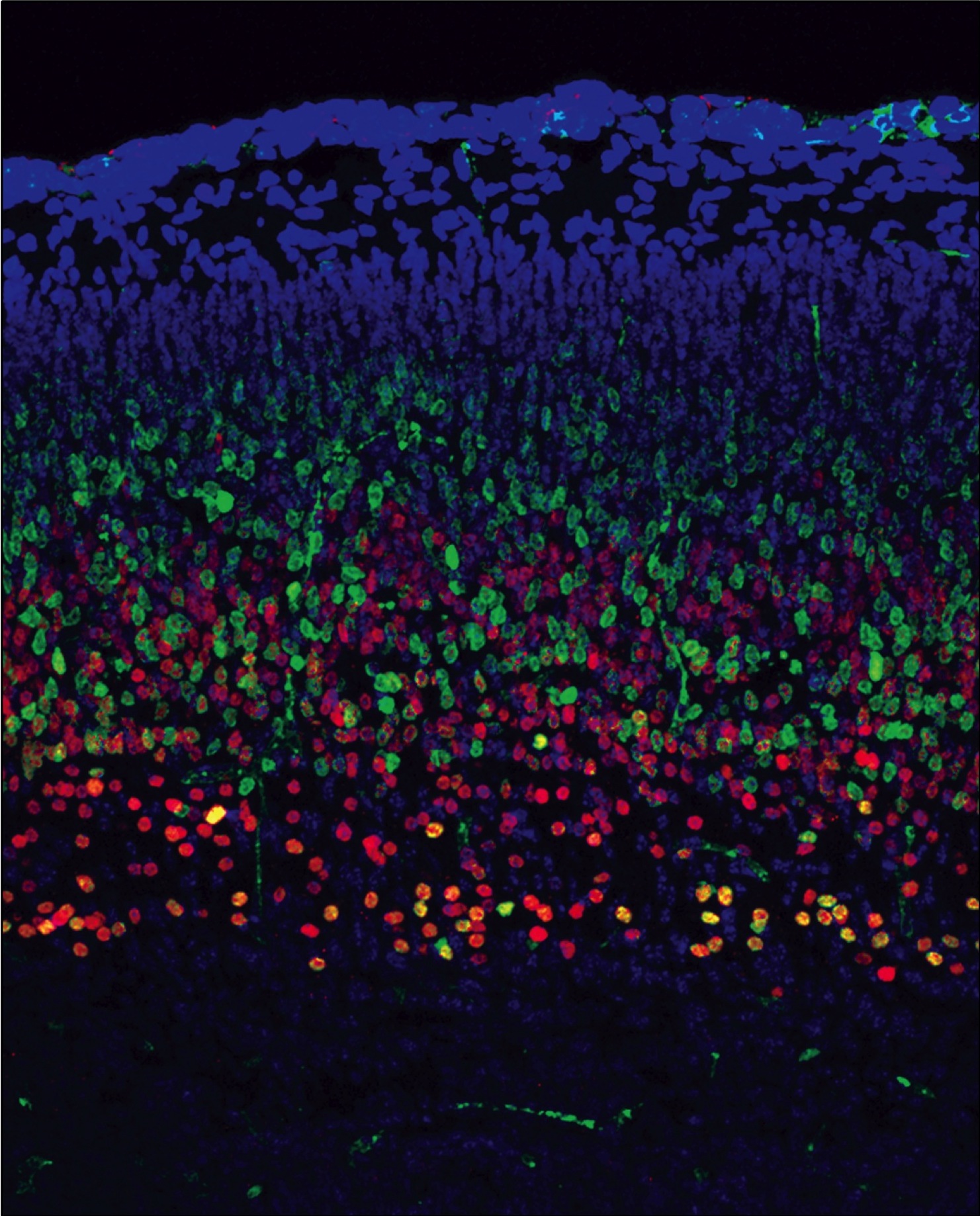
Quesnel-Vallieres, M., Irimia, M., Cordes, S.P. and Blencowe, B.J. (2015). Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes and Development. 29(7):746-759.
[PUBMED] [PDF]
|
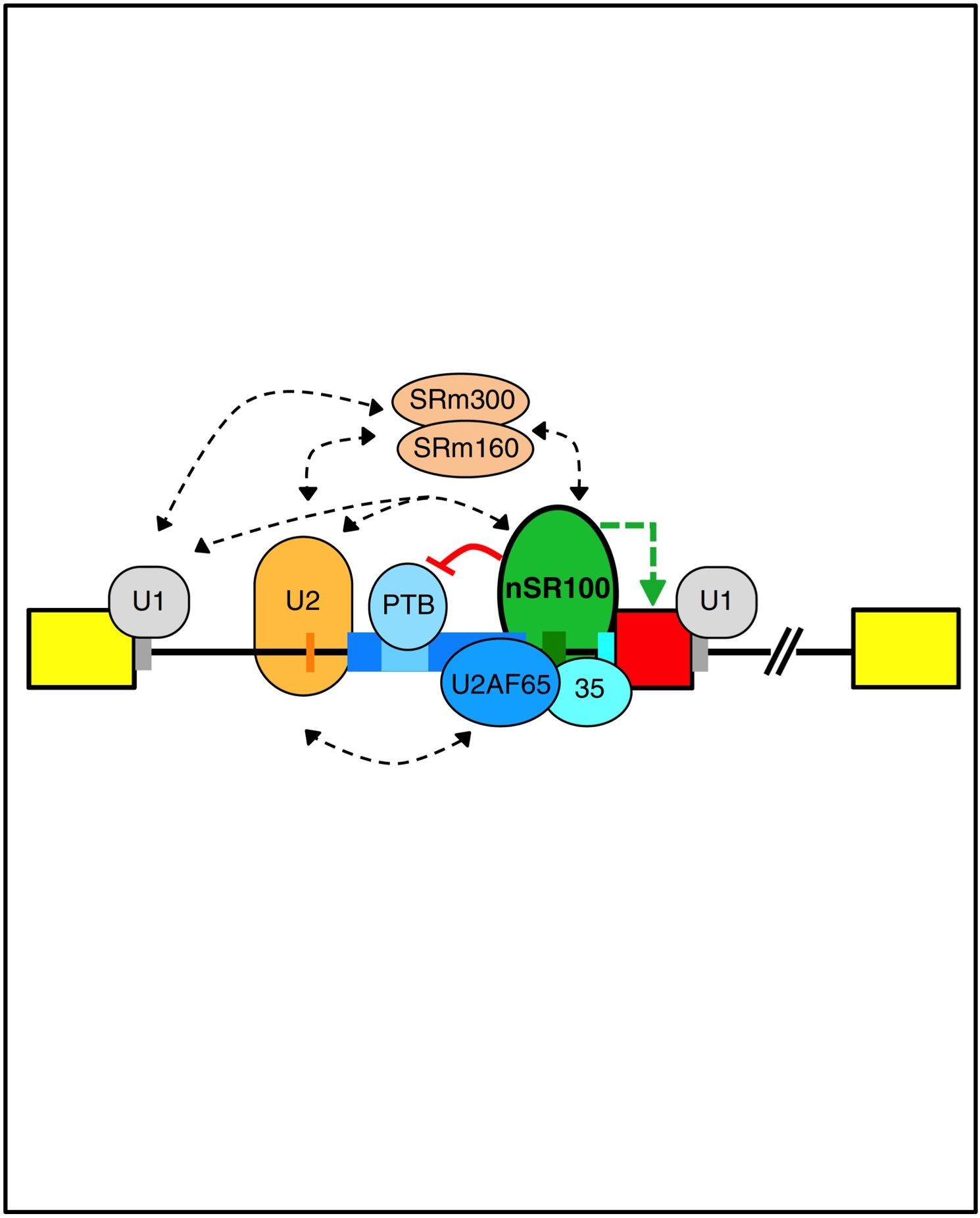
Raj, B., Irimia, M., Braunschweig, U., Sterne-Weiler, T., O’Hanlon, D., Lin, Z.Y., Chen, G.I., Easton, L., Ule, J., Gingras, AC., Eyras, E. and Blencowe, B.J. (2014). A global regulatory mechanism for activating an exon network required for neurogenesis. Molecular Cell. 56(1):90-103.
[PUBMED] [PDF]
|
Ellis, J.D., Barrios-Rodiles, M., Çolak, R., Irimia, M., Kim, T., Calarco, J.A., Wang, X., Pan, Q., O'Hanlon, D., Kim, P.M., Wrana, J.L., and Blencowe, B.J. (2012) Tissue-specific alternative splicing remodels protein-protein interaction networks. Molecular Cell. 46: 884-892.
[PUBMED] [PDF]
-Featured on the journal cover.
-Featured in Nature Reviews Genetics Editorial
|
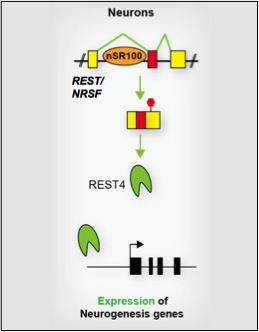
Raj, B., O'Hanlon, D., Vessey, J.P., Pan, Q., Ray, D., Buckley, N.J., Miller, F.D., and Blencowe, B.J. (2011) Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol Cell, 43: 843-850.
[PUBMED] [PDF]
|
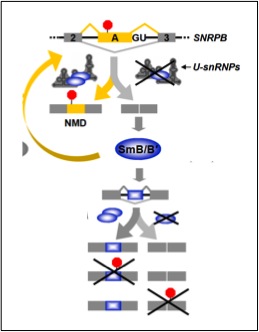
Saltzman, A.L., Pan, Q., and Blencowe, B.J. (2011). Regulation of alternative splicing by the core spliceosomal machinery. Genes and Development, 25: 373-384.
[PUBMED] [PDF]
|
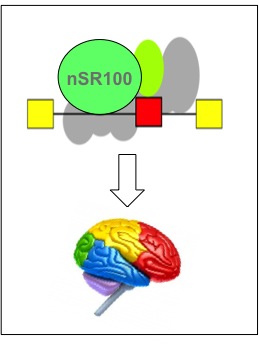
Calarco, J., Superina, S., O'Hanlon, D., Gabut, M., Raj, B., Pan, Q., Skalska, U., Clarke, L., Gelinas, D., van der Kooy, D., Zhen, M., Ciruna, B., and Blencowe, B.J. (2009). Regulation of Vertebrate Nervous System-Specific Alternative Splicing and Development by an SR-Related Protein. Cell, 138: 898-910.
[PUBMED] [PDF]
-Recommended by Faculty of 1000.
-Featured in the UofT Faculty of Medicine News.
|
Regulation of embryonic stem cell-specific alternative and splicing reprogramming
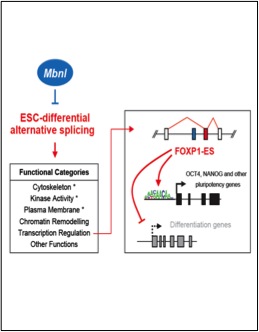
Han, H., Irimia, I., Ross, P.J., Sung, H-K., Alipanahi, B., David, L., Golipour, A., Gabut, M., Michael, I.P., Nachman E.N., Wang, E., Trcka, D., Thompson, T., O'Hanlon, D., Slobodeniuc, V., Barbosa-Morais, N.L., Burge, C.B., Moffat, J., Frey, B.J., Nagy, A., Ellis, J., Wrana, J.L. and Blencowe, B.J. (2013) MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature,
[PUBMED] [PDF]
-Highlighted in Nature News and Views article and in Nature Reviews Molecular Cell Biology article.
-Recommended by Faculty of 1000.
|
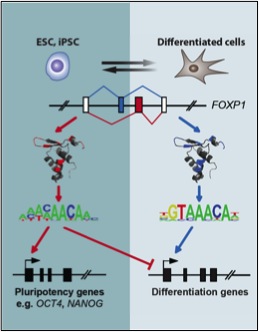
Gabut, M., Samavarchi-Tehrani, P., Wang, X., Slobodeniuc, V., O'Hanlon, D., Sung, H.K., Alvarez, M., Talukder, S., Pan, Q., Mazzoni, E.O., Nedelec, S., Wichterle, H., Woltjen, K., Hughes, T.R., Zandstra, P.W., Nagy, A., Wrana, J.L., and Blencowe, B.J. (2011) An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell, 147: 132-146.
[PUBMED] [PDF]
-Featured in Cell Preview Article and in Nature Reviews Genetics Research Highlights.
-Ranked among Top 10 "hottest" research articles in the fields of biochemistry, genetics and molecular biology in 2011
-Recommended by Faculty of 1000.
|
Role of epigenetic modifications and the transcriptional machinery in the regulation of alternative splicing
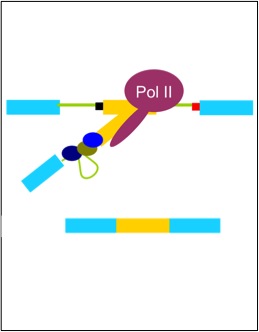
Ip, J.Y., Schmidt, D., Pan, Q., Ramani, A.K., Fraser, A.G., Odom, D.T., and Blencowe, B.J. (2011). Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Research, 21: 390-401.
[PUBMED] [PDF]
|
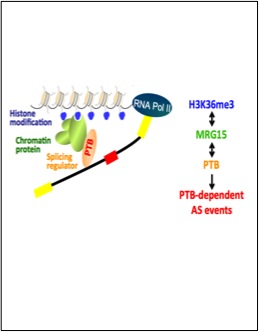
Luco, R.F., Pan, Q., Tominaga, K., Blencowe, B.J., Pereira-Smith, O.M. and Misteli, T. (2010) Regulation of alternative splicing by histone modifications. Science, 327:996-1000.
[PUBMED] [PDF]
-Recommended by Faculty of 1000.
-Featured in Dev. Cell Editorial and Nature Journal Club
|
The evolutionary landscape of alternative splicing in vertebrate species
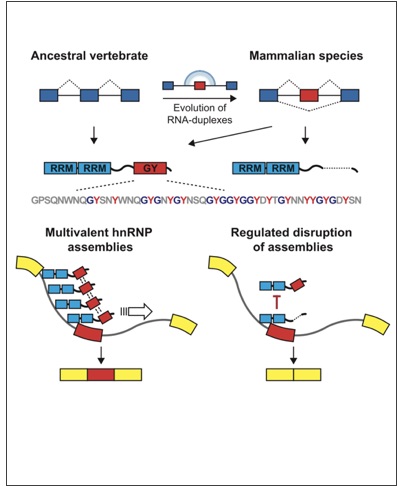
Gueroussov, S., Weatheritt, R.J., O'Hanlon, D., Lin, Z.Y., Narula, A., Gingras, A.C. and Blencowe B.J. (2017)
Regulatory Expansion in Mammals of Multivalent hnRNP Assemblies that Globally Control Alternative Splicing. Cell 170:324-339.
[PUBMED] [PDF]
- Highlighted in Nature Reviews Molecular Cell Biology
|
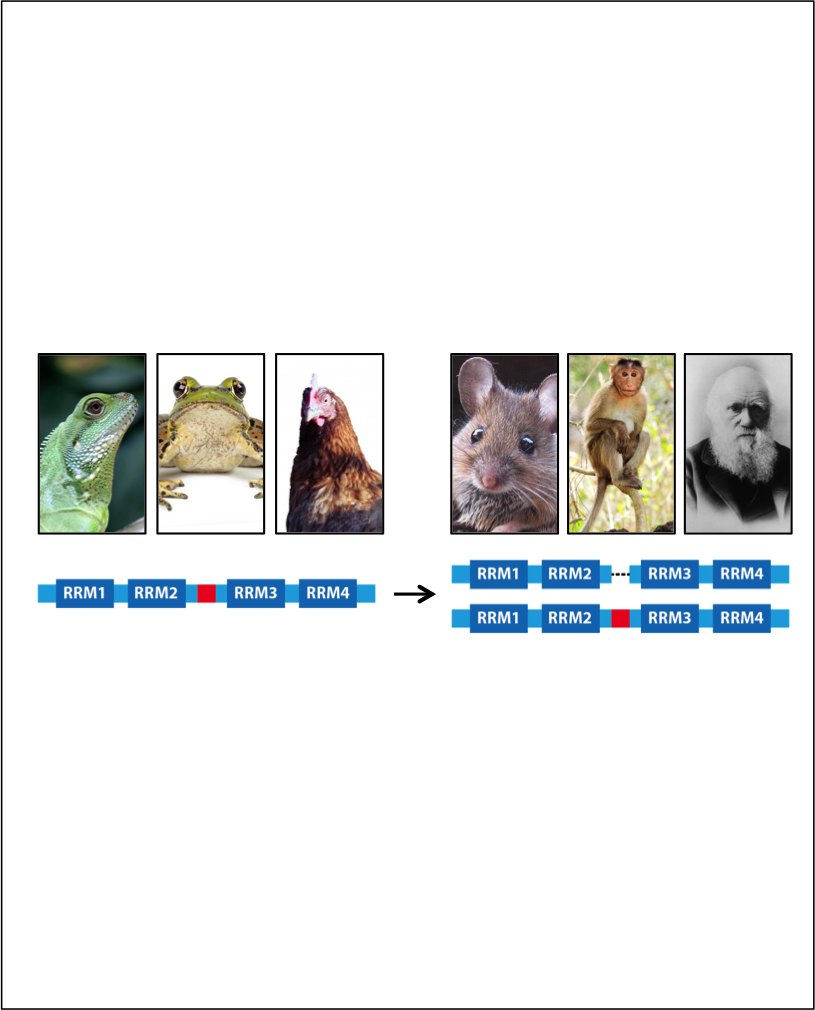
Gueroussov, S., Gonatopoulos-Pournatzis, T., Irimia, M., Raj, B., Lin, Z.Y., Gingras, A.C. and Blencowe, B.J (2015) An alternative splicing event amplifies evolutionary differences between vertebrates. Science, 349: 868-873.
[PUBMED] [PDF]
-Recommended by Faculty of 1000.
|

Barbosa-Morais, N.L., Irimia, M., Pan, Q., Xiong, H.Y., Gueroussov, S., Lee, L.J., Slobodeniuc, V., Kutter, C., Watt, S., Colak, R., Kim, T.H, Misquitta-Ali, C.M., Wilson, M.D., Kim, P.M., Odom, D.T., Frey, B.J. and Blencowe, B.J (2012) The evolutionary landscape of alternative splicing in vertebrate species, Science, 338: 1587-1593.
[PUBMED] [PDF]
-Featured in Science perspective article.
-Recommended by Faculty of 1000.
|

Calarco, J.A., Xing, Y., Caceres, M., Calarco, J.P., Xiao, X., Pan, Q., Lee, C., Preuss, T.M. and Blencowe, B.J., (2007) Global analysis of alternative splicing differences between human and chimpanzees. Genes and Dev. 21: 2963-2975.
[PUBMED] [PDF]
-Featured in CBC news and other media reports.
-Featured in Cell, Leading Edge: Genetics Select
|
Alternative splicing misregulation in human diseases and disorders

Quesnel-Vallières M., Dargaei Z., Irimia M., Gonatopoulos-Pournatzis T., Ip J., Wu M., Sterne-Weiler T., Nakagawa S., Woodin M.A., Blencowe B.J. and Cordes S.P. (2016)
Misregulation of an activity-dependent splicing network as a common mechanism underlying autism spectrum disorders. Mol. Cell 64:1023-34.
[PUBMED] [PDF]
- Featured in The Globe and Mail, The Independent, Huffington Post, The Scientist, Global News, and other media outlets.
|
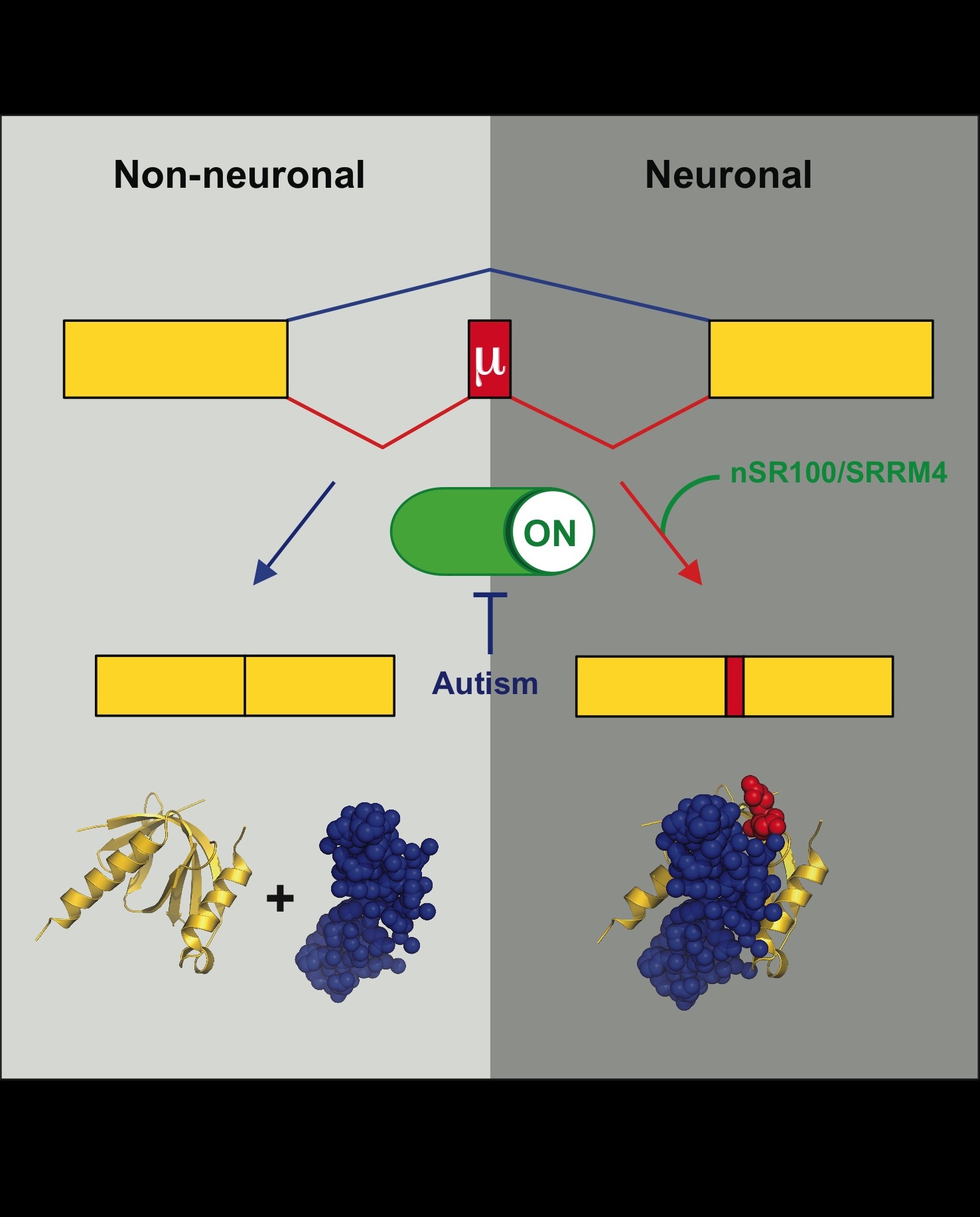
Irimia, M., Weatheritt, R.J., Ellis, J., Parikshak, N.N., Gonatopoulos-Pourtnazis, T., Babor, M., Quesnel-Vallieres, M., Tapial, J., Raj, B., O'Hanlon, D., Barrios-Rodiles, M., Sternberg, M.J.E., Cordes, S.P., Roth, F.P., Wrana, J.L., Geschwind, D.H. and Blencowe, B.J. (2014). A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 159(7):1511-1523.
[PUBMED] [PDF]
-Featured in Cell Preview, EMBO Journal Preview article and Nature Reviews Neuroscience.
-Recommended by Faculty of 1000.
-Highlighted in Toronto Star and London Free Press newspaper articles
|
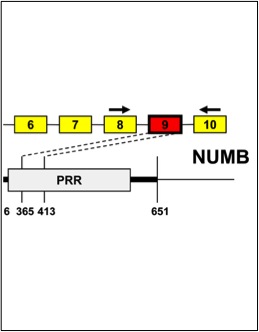
Misquitta-Ali, C.M., Cheng, E., O’Hanlon, D., Liu, N., McGlade, C.J., Tsao, M.S., and Blencowe, B.J. (2011). Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol. Cell Biol., 31: 138-150.
[PUBMED] [PDF]
|
Voineagu, I., Wang, X., Johnston, P., Lowe, J., Tian, Y., Horvath, S., Mill, J., Cantor, R., Blencowe, B.J., and Geschwind, D. (2011) Transcriptome Analysis of Autistic Brain Reveals Convergent Molecular Pathology. Nature, 474: 380-400
[PUBMED] [PDF]
-Featured in Nature, Nature Review Genetics and Nature Reviews Neuroscience Research highlights
-Recommended by Faculty of 1000.
-Highlighted in BBC, TIME and other International Media reports
|
|

































